【5.4.3.1】DNA Chisel,一种多功能序列优化器
- 网页工具: https://cuba.genomefoundry.org/sculpt_a_sequence
- 开源 Python 库使用 https://github.com/Edinburgh-Genome-Foundry/DNAChisel
- 说明文档: https://edinburgh-genome-foundry.github.io/DnaChisel/
已经提出了软件解决方案来解决各种情况,包括宿主特异性密码子优化或协调(Claassens等人,2017;Richardson等人,2012),通过CpG岛富集增强基因表达(Raab等人,2010),生物中性序列的设计(Casini等人,2014)或去除合成阻碍的DNA模式(Oberortner等人,2017)。然而,这些项目专注于特定的目标和预定的序列位置(如编码区域),并且很难集成到同一个工作流程中,因为它们的优化可能会相互抵消。D-tailor 框架(Guimaraes et al., 2014)提出了一种编程解决方案,使用户能够通过 Python 脚本自由定义和组合规范,重点是探索多目标问题。
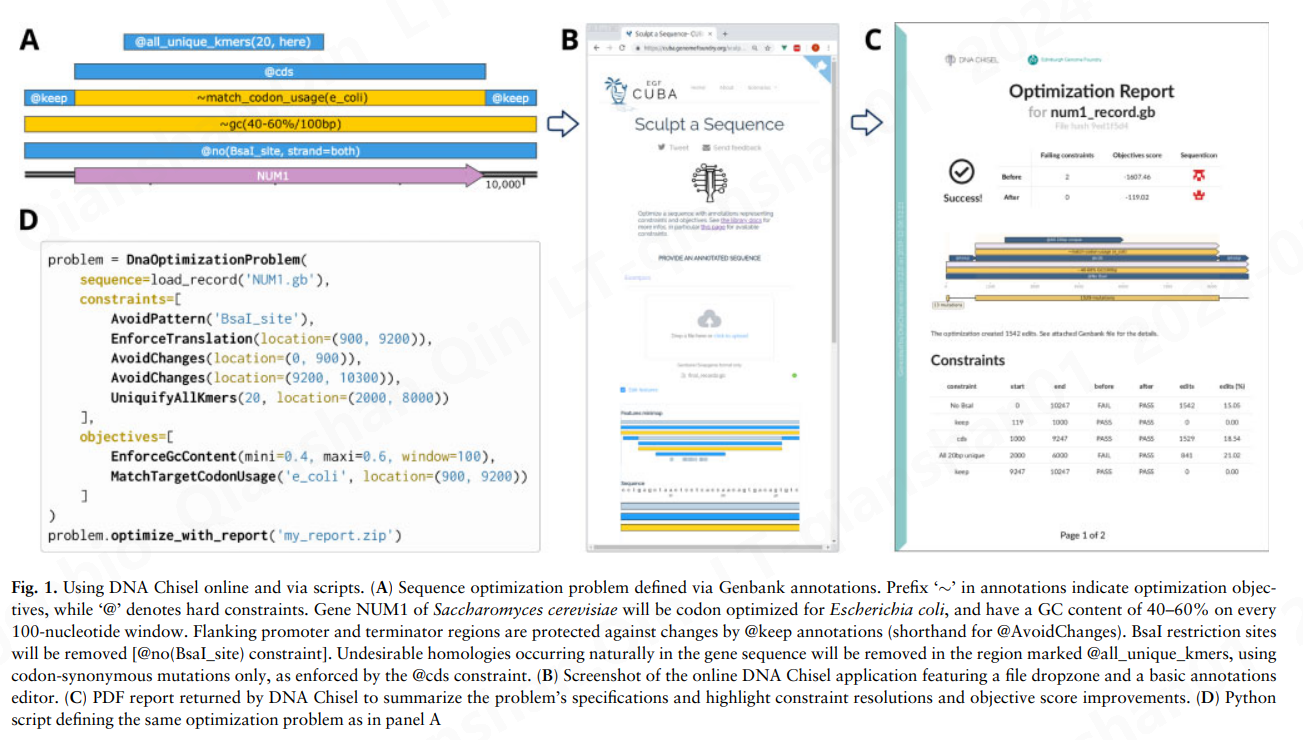
在 DNA Chisel 中,优化问题由全局或局部规范列表定义,根据这些规范将优化起始线性或循环序列。规范可以是硬约束,必须在最终序列中满足,也可以是优化目标,其分数必须最大化。例如,规范 AvoidChanges 可以用作禁止在给定区域中修改序列的约束,也可以用作简单地惩罚该区域中的更改的目标。在存在多个优化目标(可归因于相对权重)的情况下,DNA Chisel 将寻求使用下一节中描述的启发式方法最大化总加权分数(多目标基因优化的示例在补充部分S1B).
一、软件介绍
1.1 软件可以做什么
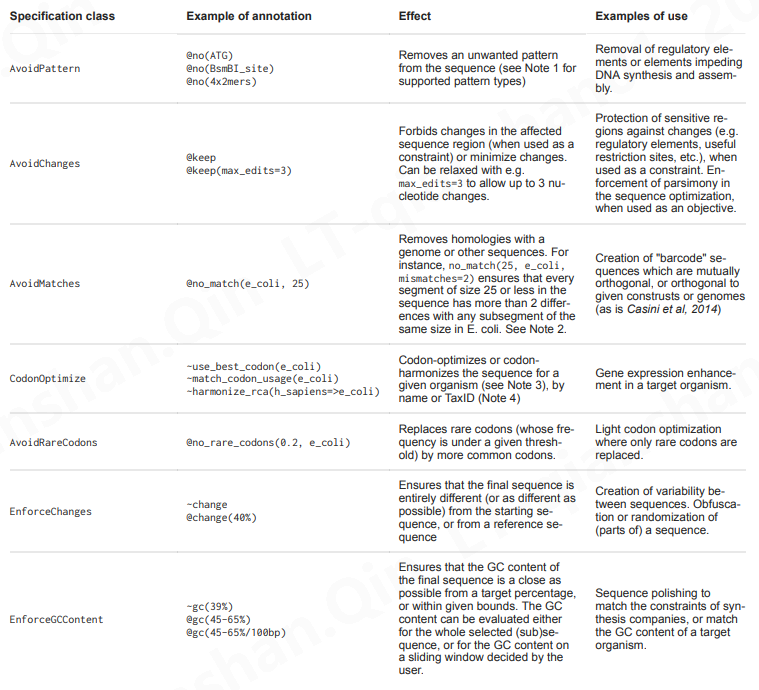
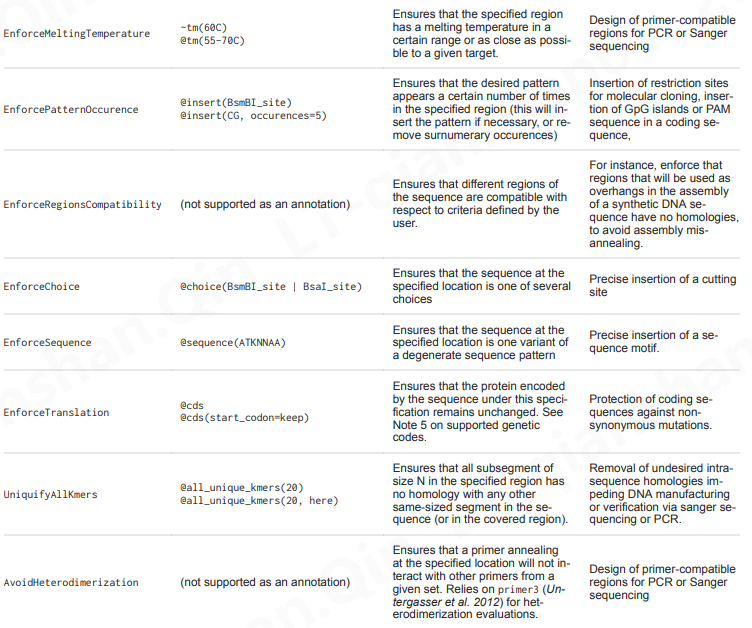
https://edinburgh-genome-foundry.github.io/DnaChisel/ref/builtin_specifications.html
1.2 软件逻辑
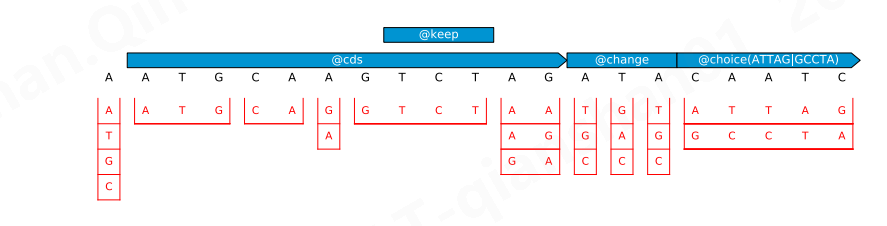
Figure S2: Restriction of the mutation space by different specification classes. In this example the problem consists of a 21-nucleotide sequence and four constraints. The mutation space, represented in red, consists of contiguous sub-segments, each associated with a set of sequence choices. For instance, the first nucleotide is unconstrained and can take any of the four possible values. The next nucleotides are constrained by @cds which enforces synonymous codons mutations, and these restrictions are combined with a @keep constraint which keeps the affected sequence segment in its original state.
优化算法
DNA Chisel 的算法首先确保验证所有约束,然后根据约束优化目标。 求解器遵循以下过程:
- 对于每个约束:
- 评估问题的约束以找到所有违规的位置。
- 对于每个违规位置,从左到右: 定义本地问题以通过本地搜索解决违规问题,同时确保不要对已经本地验证的约束造成新的破坏。
- 对于每个优化目标:
- 评估问题的目标以找到所有次优区域的位置。
- 对于每个次优区域,从左到右: 定义一个局部问题并用它来优化局部区域,以提高总体目标得分,同时确保所有约束得到验证。
虽然其他一些框架也使用局部优化,但 DNA Chisel 引入了新技术(在下一段中描述)来简化局部问题并加速解决。
定义局部问题
在DNA Chisel中,局部问题是问题的一个版本,其中只有一小段序列(在本节中表示为[start, end])会发生突变,以便局部解决特定的约束破坏,或增加序列相对于局部目标的适应度。

局部序列优化
- 选择一种搜索方法来探索突变空间。
- 使用此方法并找到满足所有约束(解决约束时)或最大化目标分数(优化目标时)的序列变体。
- 将问题的序列替换为成功的变体,然后转到下一个位置进行优化
如果序列只允许少量的变体,求解器执行穷举搜索。如果这个数字超过某个阈值(可以由用户设置,默认为10,000),则使用引导随机搜索。最后,在被解析(或优化)的规范实现其自己的解析方法的(不常见的)情况下,将使用此自定义方法
二、本地软件
说明文档: https://edinburgh-genome-foundry.github.io/DnaChisel/
2.1 安装
pip install dnachisel # <= minimal install without reports support
pip install 'dnachisel[reports]' # <= full install with all dependencies
2.2 使用
目标:
* It will be rid of BsaI sites (on both strands).
* GC content will be between 30% and 70% on every 50bp window.
* The reading frame at position 500-1400 will be codon-optimized for E. coli.
代码:
from dnachisel import *
# DEFINE THE OPTIMIZATION PROBLEM
problem = DnaOptimizationProblem(
sequence=random_dna_sequence(10000),
constraints=[
AvoidPattern("BsaI_site"),
EnforceGCContent(mini=0.3, maxi=0.7, window=50),
EnforceTranslation(location=(500, 1400))
],
objectives=[CodonOptimize(species='e_coli', location=(500, 1400))]
)
# SOLVE THE CONSTRAINTS, OPTIMIZE WITH RESPECT TO THE OBJECTIVE
problem.resolve_constraints()
problem.optimize()
# PRINT SUMMARIES TO CHECK THAT CONSTRAINTS PASS
print(problem.constraints_text_summary())
print(problem.objectives_text_summary())
# GET THE FINAL SEQUENCE (AS STRING OR ANNOTATED BIOPYTHON RECORDS)
final_sequence = problem.sequence # string
final_record = problem.to_record(with_sequence_edits=True)
输出结果:
from dnachisel import DnaOptimizationProblem
problem = DnaOptimizationProblem.from_record("my_record.gb")
problem.optimize_with_report(target="report.zip")
分析问题:
problem = DnaOptimizationProblem(...)
problem.optimize_with_report(target="report.zip")
三、我的案例
3.1 替换某个motif
from dnachisel import *
# from dnachisel.biotypes import Protein
one_seq = 'ATGAAGGCGATCATCGTCCTGCTCATGGTGGTGACGAGCAACGCGGATCGGATCTGCACCGGGATCACCTCCAGCAATTCACCTCACGTGGTG'
problem = DnaOptimizationProblem(
sequence=one_seq,
constraints=[
AvoidPattern("GTGG",strand=0),
EnforceTranslation()
],
) # objectives=[CodonOptimize(species='h_sapiens')] sequence_type=Protein,
# SOLVE THE CONSTRAINTS, OPTIMIZE WITH RESPECT TO THE OBJECTIVE
problem.resolve_constraints()
problem.optimize()
# PRINT SUMMARIES TO CHECK THAT CONSTRAINTS PASS
print(problem.constraints_text_summary())
print(problem.objectives_text_summary())
# GET THE FINAL SEQUENCE (AS STRING OR ANNOTATED BIOPYTHON RECORDS)
final_sequence = problem.sequence # string
final_record = problem.to_record(with_sequence_edits=True)
print(final_sequence)
print(final_record)
EnforceTranslation 太重要了, 保证序列翻译出来的氨基酸序列不变。。
如果Motif是一个List,可以这样
avoid_patterns = [EnforceTranslation()]
for one_mo in moitf_list:
avoid_patterns.append(AvoidPattern(one_mo, strand=0),)
problem = DnaOptimizationProblem(
sequence=one_seq,
constraints= avoid_patterns,
)
参考资料
- https://academic.oup.com/bioinformatics/article/36/16/4508/5869515?login=false 。 DNA Chisel, a versatile sequence optimizer
