【4.1.1.2】预测核酸二级结构稳定性的最近邻参数数据库--NNDB
RNA 折叠的最近邻参数(Nearest neighbor parameters)在软件包中广泛使用,但当前不存在已发布的资源来提供这些参数及其使用教程。 该数据库为社区提供此功能,并计划随着参数的完善而不断增长。 该站点还将用于提供随后完善的参数集的历史记录。
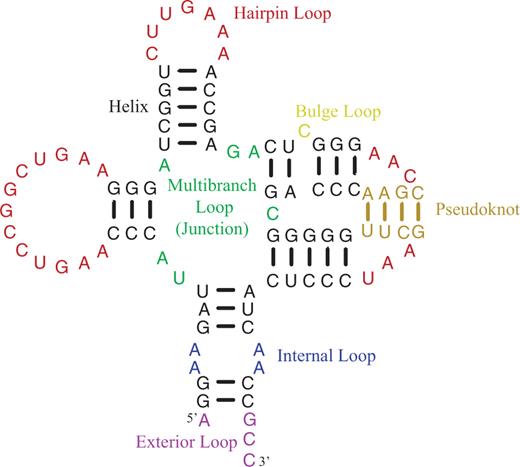
- Loops are composed of nucleotides not in canonical pairs.
- Hairpin loops have one exiting helix.
- Internal and bulge loops have two exiting helices.
- Internal loops have nucleotides not in canonical pairs on each of two strands, but bulge loops have nucleotides not in canonical pairs on only one strand.
- Multibranch loops, also called helical junctions, have three or more exiting helices.
- Exterior loops contain the ends of sequences and one or more exiting helices.
- Pseudoknots are canonical pairs connecting loop regions closed by other helices. Formally, a pseudoknot occurs when there are at least two pairs, with indices i paired to j and i′ paired to j′, that satisfy the condition i < i′ < j < j′.
- The pseudoknot helix is often considered to be composed of the fewest pairs that need to be removed to relieve the pseudoknot (19). In this structure, the tan nucleotides are in pairs that could be removed to relieve the pseudoknot.
本教程演示了使用 2004 参数预测 6 个未配对核苷酸的发夹环的折叠自由能变化
RNA(Turner 2004 ) 这些是 Turner 小组编制的当前 RNA 折叠的最近邻参数集。 37 ℃ 下的自由能变化和焓变化均已估计,从而可以预测任意温度下的结构。 参数可以文本格式或html格式下载。 提供了功能形式的描述和使用教程。
Mathews, D.H., Disney, M.D., Childs, J.L., Schroeder, S.J., Zuker, M. and Turner, D.H. (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA, 101, 7287-7292.
RNA (Turner 1999)
Mathews, D.H., Sabina, J., Zuker, M. and Turner, D.H. (1999) Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J. Mol. Biol., 288, 911-940.
DNA (RNAstructure)
Reuter, J. S., & Mathews, D. H. (2010). RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129.
RNA + m6A (Kierzek et al.)
Kierzek, E., Zhang, X., Watson, R. M., Kierzek, R., Mathews, D. H. (2022). Secondary Structure Prediction for RNA Sequences Including N6-methyladenosine. Nature Communications, 13, 1271.
https://rna.urmc.rochester.edu/NNDB/
参考资料
- Douglas H. Turner, David H. Mathews, NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure, Nucleic Acids Research, Volume 38, Issue suppl_1, 1 January 2010, Pages D280–D282, https://doi.org/10.1093/nar/gkp892
- NNDB: An expanded database of nearest neighbor parameters for predicting stability of nucleic acid secondary structures . https://doi.org/10.1016/j.jmb.2024.168549
- Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. https://doi.org/10.1006/jmbi.1999.2700
这里是一个广告位,,感兴趣的都可以发邮件聊聊:tiehan@sina.cn
![]() 个人公众号,比较懒,很少更新,可以在上面提问题,如果回复不及时,可发邮件给我: tiehan@sina.cn
个人公众号,比较懒,很少更新,可以在上面提问题,如果回复不及时,可发邮件给我: tiehan@sina.cn

