【9.1.5】5-prime capping of mRNA
The cap structure at the 5′ end of mRNA is critical for the efficient translation, stabilization, and transportation of mRNAs in eukaryotes. RNA capping is an essential part of the workflow when mRNA synthesized in vitro (in vitro-transcribed or IVT mRNA) for downstream applications related to translation.
Basic mRNA structure
Figure 1 shows the basic structure of in vitro-transcribed mRNA, which includes a 5′ cap, a 5′ untranslated region (UTR), a coding sequence (CDS) containing start and stop codons, a 3′ UTR, and a poly(A) tail. These mRNA features are all necessary to create a loop structure via poly(A) binding protein (PABP), enabling efficient translation. A modified nucleoside, such as pseudouridine is often used to suppress immunogenicity for the host cells (Kariko et al., 2008).
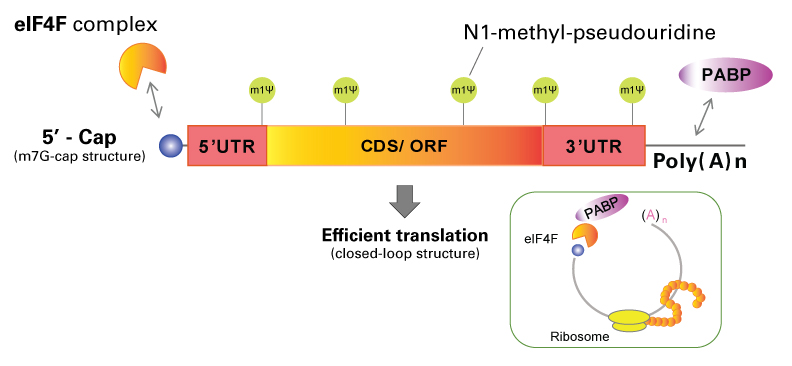
Figure 1. In vitro-transcribed mRNA structure. eIF4F = Eukaryotic initiation factor 4F, 5′ UTR = 5′ untranslated region, CDS/ORF = coding sequence/open reading frame, 3′ UTR = 3′ untranslated region, PABP = poly(A) binding protein, m1Ψ = N1-methylpseudouridine.
The 5′ cap is guanosine with a methyl group on the 7-position; this modified nucleoside is often abbreviated as m7G. The cap connects via triphosphate linkage (PPP) to the first nucleotide at the 5′ end of mRNA, where transcription is initiated (the +1 position).
The initial 5′ cap structure (m7G-ppp-N1) in mRNA is referred to as a cap-0 structure. In eukaryotes, the first nucleotide (+1) adjacent to the 5′ cap can be further modified to create a cap-1 structure, which suppresses immunogenicity.
Cap-0 and cap-1 mRNA structures
A 5′ cap-0 structure occurs when there is no modification to the adjacent nucleotide. Adding a methyl group to the first nucleotide creates a cap-1 structure. Cap-0 and cap-1 structures are shown in Figure 2. Since the cap-1 structure is involved in distinguishing self from nonself, it triggers less immunogenicity when administered in vivo.
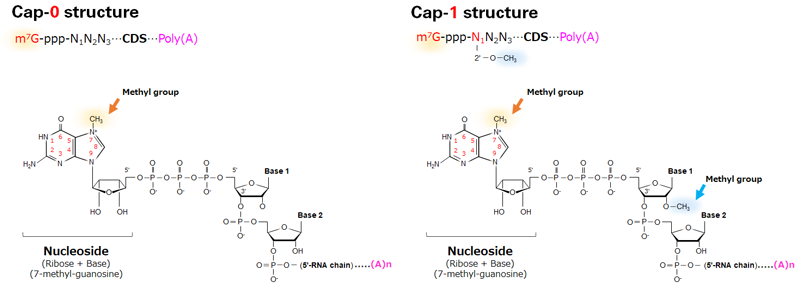
Figure 2. Comparison of cap-0 and cap-1 structures. Cap-1 mRNA has an additional methyl group within the first nucleotide, as indicated by the blue arrow on the right. m7G = 5′ cap, ppp = triphosphate linkage, N1N2N3 = transcription start/initiation site (the first 3 nucleotides of the 5′ UTR), CDS = coding sequence.
Co-transcriptional vs. post-transcriptional capping
There are two major ways to generate cap-1 mRNAs. The first is co-transcriptional capping using cap analogs (Figure 3, left panel). The second is post-transcriptional capping using enzymes such as Vaccinia Capping Enzyme (VCE, Cat. #2460) and mRNA Cap 2′-O-Methyltransferase (2′-O-MTase, Cat. #2470) as shown in Figure 3, right panel.

Figure 3. Co-transcriptional vs. post-transcriptional approaches to creating the cap-1 structure. Co-transcriptional capping is performed by adding cap analogs such as CleanCap® (TriLink BioTechnologies) to the 5′-end of newly synthesized mRNA during the in vitro transcription reaction. In post-transcriptional capping, the 5′ cap (cap-0 structure) is first generated by vaccinia capping enzyme (VCE). Then, the transcription start site (N1) is modified by mRNA cap 2'-O-methyltransferase (2'-O-MTase) to further generate a cap-1 structure.
参考资料
