【4.1.1】模型选择与评估--1--交叉检验(Cross Validation)
一、交叉验证概述
进行模型验证的一个重要目的是要选出一个最合适的模型,对于监督学习而言,我们希望模型对于未知数据的泛化能力强,所以就需要模型验证这一过程来体现不同的模型对于未知数据的表现效果。
最先我们用训练准确度(用全部数据进行训练和测试)来衡量模型的表现,这种方法会导致模型过拟合;为了解决这一问题,我们将所有数据分成训练集和测试集两部分,我们用训练集进行模型训练,得到的模型再用测试集来衡量模型的预测表现能力,这种度量方式叫测试准确度,这种方式可以有效避免过拟合。
测试准确度的一个缺点是其样本准确度是一个高方差估计(high varianceestimate),所以该样本准确度会依赖不同的测试集,其表现效果不尽相同。
cross-validation (CV for short)
二、K折交叉验证
- 将数据集平均分割成K个等份
- 使用1份数据作为测试数据,其余作为训练数据
- 计算测试准确率
- 使用不同的测试集,重复2、3步骤
- 对测试准确率做平均,作为对未知数据预测准确率的估计
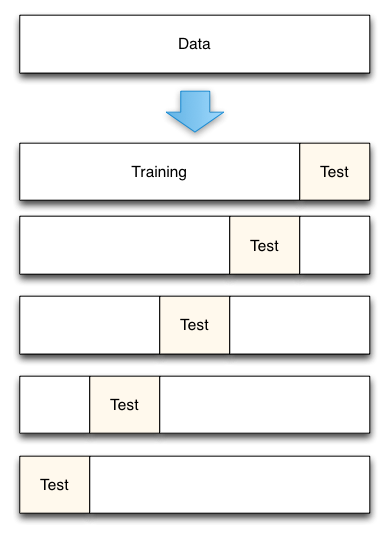
案例如下:
sklearn 0.20以后的代码:
>>> from sklearn.model_selection import KFold
>>> X = np.array([[1, 2], [3, 4], [1, 2], [3, 4]])
>>> y = np.array([1, 2, 3, 4])
>>> kf = KFold(n_splits=2)
>>> kf.get_n_splits(X)
2
>>> print(kf)
KFold(n_splits=2, random_state=None, shuffle=False)
>>> for train_index, test_index in kf.split(X):
... print("TRAIN:", train_index, "TEST:", test_index)
... X_train, X_test = X[train_index], X[test_index]
... y_train, y_test = y[train_index], y[test_index]
TRAIN: [2 3] TEST: [0 1]
TRAIN: [0 1] TEST: [2 3]
方法说明:
get_n_splits([X, y, groups]) Returns the number of splitting iterations in the cross-validator
split(X[, y, groups]) Generate indices to split data into training and test set.
参数说明:
sklearn.model_selection.KFold(n_splits=3, shuffle=False, random_state=None)
- n_splits : 默认3,最小为2;拆成几分 shuffle : boolean, 默认False;shuffle会对数据产生随机搅动(洗牌) random_state : int, 默认None,随机种子
如果想对数据分层呢??
sklearn 0.20以前的代码:
from sklearn.cross_validation import KFold
input_seqs = range(25)
kf = KFold(len(input_seqs), n_folds=5)
print input_seqs
#[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24]
for train_index, test_index in kf:
train_seqs, test_seqs = np.array(input_seqs)[train_index], np.array(input_seqs)[test_index]
print train_seqs,test_seqs
结果如下:
[ 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24] [0 1 2 3 4]
[ 0 1 2 3 4 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24] [5 6 7 8 9]
[ 0 1 2 3 4 5 6 7 8 9 15 16 17 18 19 20 21 22 23 24] [10 11 12 13 14]
[ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 20 21 22 23 24] [15 16 17 18 19]
[ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19] [20 21 22 23 24]
函数解释:
sklearn.cross_validation.KFold(n, n_folds=3, shuffle=False, random_state=None)
参数:
n: 代表数据的个数
n_splits : 默认3,最小为2;K折验证的K值
shuffle : 默认False;shuffle会对数据产生随机搅动(洗牌)
random_state :默认None,随机种子
方法:
get_n_splits([X, y, groups]) Returnsthe number of splitting iterations in the cross-validator
split(X[, y, groups]) Generateindices to split data into training and test set.
上面例子分出25个数据的五分之一作为测试集,剩下的作为训练集,这里没有对数据进行洗牌,所以按数据的顺序进行选择。训练-测试的组合也为五次。
三、Computing cross-validated metrics
cross validation大概的意思是:对于原始数据我们要将其一部分分为traindata,一部分分为test data。train data用于训练,test data用于测试准确率。在test data上测试的结果叫做validation error。将一个算法作用于一个原始数据,我们不可能只做出随机的划分一次train和testdata,然后得到一个validation error,就作为衡量这个算法好坏的标准。因为这样存在偶然性。我们必须多次的随机的划分train data和test data,分别在其上面算出各自的validation error。这样就有一组validationerror,根据这一组validationerror,就可以较好的准确的衡量算法的好坏。crossvalidation是在数据量有限的情况下的非常好的一个evaluate performance的方法。而对原始数据划分出train data和testdata的方法有很多种,这也就造成了cross validation的方法有很多种。
sklearn.cross_validation.cross_val_score(estimator, X, y=None, scoring=None, cv=None,n_jobs=1, verbose=0, fit_params=None, pre_dispatch=‘2*n_jobs’)
- estimator:是不同的分类器,可以是任何的分类器。比如支持向量机分类器:estimator = svm.SVC(kernel=‘linear’, C=1)
- cv:代表不同的cross validation的方法。如果cv是一个int值,并且如果提供了rawtarget参数,那么就代表使用StratifiedKFold分类方式;如果cv是一个int值,并且没有提供rawtarget参数,那么就代表使用KFold分类方式;也可以给定它一个CV迭代策略生成器,指定不同的CV方法。
- scoring:默认Nnoe,准确率的算法,可以通过score_func参数指定;如果不指定的话,是用estimator默认自带的准确率算法。 scoring=’f1_macro’进行评测的
例子:
>>> from sklearn.model_selection import cross_val_score
>>> clf = svm.SVC(kernel='linear', C=1)
>>> scores = cross_val_score(clf, iris.data, iris.target, cv=5)
>>> scores
array([ 0.96..., 1. ..., 0.96..., 0.96..., 1. ])
平均得分与95%的置信区间
>>> print("Accuracy: %0.2f (+/- %0.2f)" % (scores.mean(), scores.std() * 2))
Accuracy: 0.98 (+/- 0.03)
the score computed at each CV iteration is the score method of the estimator,可以通过scoring来定义方法。(不同方法代表的意思见http://scikit-learn.org/stable/modules/model_evaluation.html#scoring-parameter)
>>> from sklearn import metrics
>>> scores = cross_val_score(
... clf, iris.data, iris.target, cv=5, scoring='f1_macro')
>>> scores
array([ 0.96..., 1. ..., 0.96..., 0.96..., 1. ])
也可以通过其他的方法:
>>> from sklearn.model_selection import ShuffleSplit
>>> n_samples = iris.data.shape[0]
>>> cv = ShuffleSplit(n_splits=3, test_size=0.3, random_state=0)
>>> cross_val_score(clf, iris.data, iris.target, cv=cv)
...
array([ 0.97..., 0.97..., 1. ])
其他例子:
>>> from sklearn import preprocessing
>>> X_train, X_test, y_train, y_test = train_test_split(
... iris.data, iris.target, test_size=0.4, random_state=0) ## 快速打乱数据,这里是按照6:4对训练集测试集进行划分
>>> scaler = preprocessing.StandardScaler().fit(X_train)
>>> X_train_transformed = scaler.transform(X_train)
>>> clf = svm.SVC(C=1).fit(X_train_transformed, y_train)
>>> X_test_transformed = scaler.transform(X_test)
>>> clf.score(X_test_transformed, y_test)
0.9333...
更简单的方法:
>>> from sklearn.pipeline import make_pipeline
>>> clf = make_pipeline(preprocessing.StandardScaler(), svm.SVC(C=1))
>>> cross_val_score(clf, iris.data, iris.target, cv=cv)
...
array([ 0.97..., 0.93..., 0.95...])
The cross_validate function and multiple metric evaluation
The cross_validate function differs from cross_val_score in two ways -
- It allows specifying multiple metrics for evaluation.
- It returns a dict containing training scores, fit-times and score-times in addition to the test score.
The multiple metrics can be specified either as a list, tuple or set of predefined scorer names:
>>> from sklearn.model_selection import cross_validate
>>> from sklearn.metrics import recall_score
>>> scoring = ['precision_macro', 'recall_macro']
>>> clf = svm.SVC(kernel='linear', C=1, random_state=0)
>>> scores = cross_validate(clf, iris.data, iris.target, scoring=scoring,
... cv=5, return_train_score=False)
>>> sorted(scores.keys())
['fit_time', 'score_time', 'test_precision_macro', 'test_recall_macro']
>>> scores['test_recall_macro']
array([ 0.96..., 1. ..., 0.96..., 0.96..., 1. ])
Or as a dict mapping scorer name to a predefined or custom scoring function:
>>> from sklearn.metrics.scorer import make_scorer
>>> scoring = {'prec_macro': 'precision_macro',
... 'rec_micro': make_scorer(recall_score, average='macro')}
>>> scores = cross_validate(clf, iris.data, iris.target, scoring=scoring,
... cv=5, return_train_score=True)
>>> sorted(scores.keys())
['fit_time', 'score_time', 'test_prec_macro', 'test_rec_micro',
'train_prec_macro', 'train_rec_micro']
>>> scores['train_rec_micro']
array([ 0.97..., 0.97..., 0.99..., 0.98..., 0.98...
Here is an example of cross_validate using a single metric:
>>> scores = cross_validate(clf, iris.data, iris.target,
... scoring='precision_macro')
>>> sorted(scores.keys())
['fit_time', 'score_time', 'test_score', 'train_score']
cross_val_predict
cross_val_predict 与cross_val_score 很相像,不过不同于返回的是评测效果,cross_val_predict返回的是estimator 的分类结果(或回归值),这个对于后期模型的改善很重要,可以通过该预测输出对比实际目标值,准确定位到预测出错的地方,为我们参数优化及问题排查十分的重要。
>>> from sklearn.model_selection import cross_val_predict
>>> predicted = cross_val_predict(clf, iris.data, iris.target, cv=10)
>>> metrics.accuracy_score(iris.target, predicted)
0.973...
四、不同交叉验证(Cross validation)的生成器
. If one knows that the samples have been generated using a time-dependent process, it’s safer to use a time-series aware cross-validation scheme Similarly if we know that the generative process has a group structure (samples from collected from different subjects, experiments, measurement devices) it safer to use group-wise cross-validation.
4.1 Cross-validation iterators for i.i.d. data (独立且无规律的一堆数据)
Assuming that some data is Independent and Identically Distributed (i.i.d.)
4.1.1 K-fold
如上面示意图所示:
>>> import numpy as np
>>> from sklearn.model_selection import KFold
>>> X = ["a", "b", "c", "d"]
>>> kf = KFold(n_splits=2)
>>> for train, test in kf.split(X):
... print("%s %s" % (train, test))
[2 3] [0 1]
[0 1] [2 3]
可以通过np.array的使用,来 提取数据
>>> X = np.array([[0., 0.], [1., 1.], [-1., -1.], [2., 2.]])
>>> y = np.array([0, 1, 0, 1])
>>> X_train, X_test, y_train, y_test = X[train], X[test], y[train], y[test]
4.1.2 Repeated K-Fold
RepeatedKFold即将K-Fold重复n次
>>> import numpy as np
>>> from sklearn.model_selection import RepeatedKFold
>>> X = np.array([[1, 2], [3, 4], [1, 2], [3, 4]])
>>> random_state = 12883823
>>> rkf = RepeatedKFold(n_splits=2, n_repeats=2, random_state=random_state)
>>> for train, test in rkf.split(X):
... print("%s %s" % (train, test))
...
[2 3] [0 1]
[0 1] [2 3]
[0 2] [1 3]
[1 3] [0 2]
4.1.3 Leave One Out (LOO)
LeaveOneOut (or LOO)仅仅将数据集中的一个数据拿出来用来测试,比如有n个数据,那就可以训练n次,每次训练集是n-1个数据,测试的为1个数据。
>>> from sklearn.model_selection import LeaveOneOut
>>> X = [1, 2, 3, 4]
>>> loo = LeaveOneOut()
>>> for train, test in loo.split(X):
... print("%s %s" % (train, test))
[1 2 3] [0]
[0 2 3] [1]
[0 1 3] [2]
[0 1 2] [3]
LOO因为是用n-1个数据来训练,相当于拿整个数据集n来做训练了。因此他的high variance as an estimator for the test error。 一般建议用5- 或者10- fold的方法来分数据。(但, if the learning curve is steep for the training size in question, then 5- or 10- fold cross validation can overestimate the generalization error.)
4.1.4 Leave P Out (LPO)
LeavePOut相当于从n个数据集里拿出p个数据作为测试。因此测试的数据的可能性为 Cnp
>>> from sklearn.model_selection import LeavePOut
>>> X = np.ones(4)
>>> lpo = LeavePOut(p=2)
>>> for train, test in lpo.split(X):
... print("%s %s" % (train, test))
[2 3] [0 1]
[1 3] [0 2]
[1 2] [0 3]
[0 3] [1 2]
[0 2] [1 3]
[0 1] [2 3]
4.1.5 Random permutations cross-validation a.k.a. Shuffle & Split
自定义训练和测试数据
>>> from sklearn.model_selection import ShuffleSplit
>>> X = np.arange(5)
>>> ss = ShuffleSplit(n_splits=3, test_size=0.25,
... random_state=0)
>>> for train_index, test_index in ss.split(X):
... print("%s %s" % (train_index, test_index))
...
[1 3 4] [2 0]
[1 4 3] [0 2]
[4 0 2] [1 3]
sklearn.model_selection.ShuffleSplit(n_splits=10, test_size=0.1, train_size=None, random_state=None
- n_splits 拆分成几份
- test_size test数据集占的比重,在0-1之间。如果为整数,则代表测试数据的个数
- train_size 同理test_size
- random_state 随机的种子
4.2 Cross-validation iterators with stratification based on class labels(带标签的数据)
有些数据是带有分类标签哒
4.2.1 Stratified k-fold
StratifiedKFold 在进行kFold分类的时候,会考虑数据集带有的标签
>>> from sklearn.model_selection import StratifiedKFold
>>> X = np.ones(10)
>>> y = [0, 0, 0, 0, 1, 1, 1, 1, 1, 1]
>>> skf = StratifiedKFold(n_splits=3)
>>> for train, test in skf.split(X, y):
... print("%s %s" % (train, test))
[2 3 6 7 8 9] [0 1 4 5]
[0 1 3 4 5 8 9] [2 6 7]
[0 1 2 4 5 6 7] [3 8 9]
4.2.2 Stratified Shuffle Split
StratifiedShuffleSplit 同理ShuffleSplit,需要考虑数据的标签
>>> from sklearn.model_selection import StratifiedShuffleSplit
>>> X = np.array([[1, 2], [3, 4], [1, 2], [3, 4]])
>>> y = np.array([0, 0, 1, 1])
>>> sss = StratifiedShuffleSplit(n_splits=3, test_size=0.5, random_state=0)
>>> sss.get_n_splits(X, y)
3
>>> print(sss)
StratifiedShuffleSplit(n_splits=3, random_state=0, ...)
>>> for train_index, test_index in sss.split(X, y):
... print("TRAIN:", train_index, "TEST:", test_index)
... X_train, X_test = X[train_index], X[test_index]
... y_train, y_test = y[train_index], y[test_index]
TRAIN: [1 2] TEST: [3 0]
TRAIN: [0 2] TEST: [1 3]
TRAIN: [0 2] TEST: [3 1]
4.3 Cross-validation iterators for grouped data(分组数据)
4.3.1 Group k-fold
GroupKFold 在保证kFold的基础上,保证同一个group的数据不会拆分到train和test中。
>>> from sklearn.model_selection import GroupKFold
>>> X = [0.1, 0.2, 2.2, 2.4, 2.3, 4.55, 5.8, 8.8, 9, 10]
>>> y = ["a", "b", "b", "b", "c", "c", "c", "d", "d", "d"]
>>> groups = [1, 1, 1, 2, 2, 2, 3, 3, 3, 3]
>>> gkf = GroupKFold(n_splits=3)
>>> for train, test in gkf.split(X, y, groups=groups):
... print("%s %s" % (train, test))
[0 1 2 3 4 5] [6 7 8 9]
[0 1 2 6 7 8 9] [3 4 5]
[3 4 5 6 7 8 9] [0 1 2]
4.3.2 Leave One Group Out
LeaveOneGroupOut就是丢一个group的数据作为测试数据
>>> from sklearn.model_selection import LeaveOneGroupOut
>>> X = [1, 5, 10, 50, 60, 70, 80]
>>> y = [0, 1, 1, 2, 2, 2, 2]
>>> groups = [1, 1, 2, 2, 3, 3, 3]
>>> logo = LeaveOneGroupOut()
>>> for train, test in logo.split(X, y, groups=groups):
... print("%s %s" % (train, test))
[2 3 4 5 6] [0 1]
[0 1 4 5 6] [2 3]
[0 1 2 3] [4 5 6]
4.3.3 Leave P Groups Out
丢n个gropu来作为测试
>>> from sklearn.model_selection import LeavePGroupsOut
>>> X = np.arange(6)
>>> y = [1, 1, 1, 2, 2, 2]
>>> groups = [1, 1, 2, 2, 3, 3]
>>> lpgo = LeavePGroupsOut(n_groups=2)
>>> for train, test in lpgo.split(X, y, groups=groups):
... print("%s %s" % (train, test))
[4 5] [0 1 2 3]
[2 3] [0 1 4 5]
[0 1] [2 3 4 5]
4.3.4 Group Shuffle Split
GroupShuffleSplit 是 ShuffleSplit 和LeavePGroupsOut综合
>>> from sklearn.model_selection import GroupShuffleSplit
>>> X = [0.1, 0.2, 2.2, 2.4, 2.3, 4.55, 5.8, 0.001]
>>> y = ["a", "b", "b", "b", "c", "c", "c", "a"]
>>> groups = [1, 1, 2, 2, 3, 3, 4, 4]
>>> gss = GroupShuffleSplit(n_splits=4, test_size=0.5, random_state=0)
>>> for train, test in gss.split(X, y, groups=groups):
... print("%s %s" % (train, test))
...
[0 1 2 3] [4 5 6 7]
[2 3 6 7] [0 1 4 5]
[2 3 4 5] [0 1 6 7]
[4 5 6 7] [0 1 2 3]
4.4 Predefined Fold-Splits / Validation-Sets (已确定的测试数据)
已经名明确test_fold的情况下
>>> from sklearn.model_selection import PredefinedSplit
>>> X = np.array([[1, 2], [3, 4], [1, 2], [3, 4]])
>>> y = np.array([0, 0, 1, 1])
>>> test_fold = [0, 1, -1, 1]
>>> ps = PredefinedSplit(test_fold)
>>> ps.get_n_splits()
2
>>> print(ps)
PredefinedSplit(test_fold=array([ 0, 1, -1, 1]))
>>> for train_index, test_index in ps.split():
... print("TRAIN:", train_index, "TEST:", test_index)
... X_train, X_test = X[train_index], X[test_index]
... y_train, y_test = y[train_index], y[test_index]
TRAIN: [1 2 3] TEST: [0]
TRAIN: [0 2] TEST: [1 3]
4.5 Cross validation of time series data (时间序列数据)
4.5.1 Time Series Split
TimeSeriesSplit
>>> from sklearn.model_selection import TimeSeriesSplit
>>> X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
>>> y = np.array([1, 2, 3, 4, 5, 6])
>>> tscv = TimeSeriesSplit(n_splits=3)
>>> print(tscv)
TimeSeriesSplit(max_train_size=None, n_splits=3)
>>> for train, test in tscv.split(X):
... print("%s %s" % (train, test))
[0 1 2] [3]
[0 1 2 3] [4]
[0 1 2 3 4] [5]
参考资料:
- http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.KFold.html#sklearn.model_selection.KFold
- http://scikit-learn.org/stable/modules/generated/sklearn.cross_validation.KFold.html
- https://blog.csdn.net/cherdw/article/details/54986863
- http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.ShuffleSplit.html#sklearn.model_selection.ShuffleSplit
- http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.PredefinedSplit.html#sklearn.model_selection.PredefinedSplit
